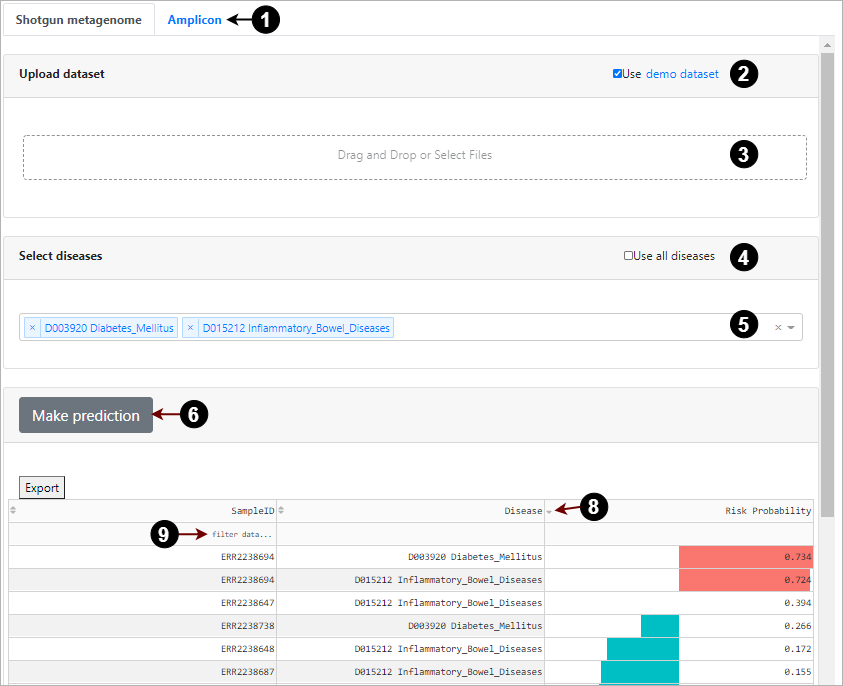
How to predict diseases risks?
As the disease prediction models are built on the human gut microbiota at the species level with the species (features) represented by NCBI taxonomic IDs. Only the species relative abundance files with rows as NCBI taxonomic IDs and columns as samples in tsv or csv format are supported. NCBI taxonomy database is recommended for taxonomic classification information assignment.
Select how your sequences generated (1), shotgun metagenome or amplicon sequencing, on the ‘Risk Prediction’ page. Upload your species relative abundance file (3) or use the demo dataset (2) for an example. Select diseases (5) you want to predict the risk or use all of the diseases (4) available in GutBalance. Press the ‘Make prediction’ button (6) to run the prediction process, and the predicted results will show in the output panel. The output panel supports the interactive exploration for searching, filtering (9), or sorting (8) by the sample id, disease id, and risk scores, according to specific requirements.
How to infer new microbe-disease associations based on GBDAD?
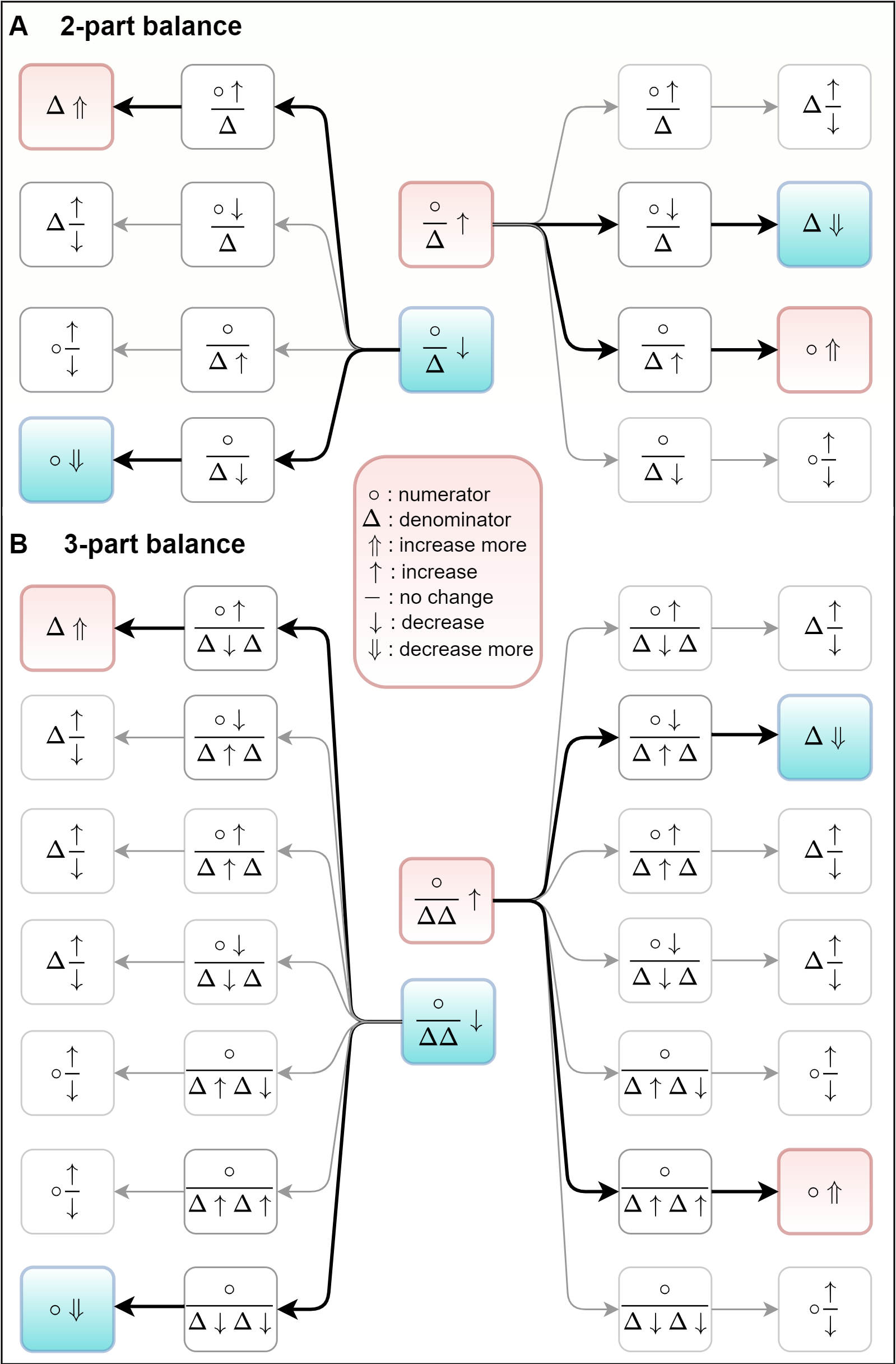
Based on the schematic diagram of all of the possible scenarios about the absolute changes of microbes that may happen when the 2-part (A) or 3-part (B) balances were identified to be positively (red color) or negatively (blue color) related to a disease phenotype. Specifically, there are eight scenarios from which the new taxon-disease association can be uniquely inferred are outlined in colored boxes.
For example, suppose a 2-part balance (A) is positively correlated (increased in disease samples, colored red) with a disease phenotype, and the abundance of the taxon in the denominator is known to increase in the respective phenotype (positively correlated with the disease). Thus, we can infer the abundance of the taxon in the numerator is raising in this phenotype and raising more than the taxon in the denominator.
The evidenced species-disease associations can be retrieved in the GBDAD database facilitating the generation of new taxon-disease association hypothesis based on the change of balances.
The credibility of the new species-disease associations inferred from GBDAD is directly influenced by the reliability of the predicted balance-disease associations and the evidenced microbe-disease associations. Experimental validation or prior biological knowledge will be required to confirm the inference further.
The case study of GBDAD
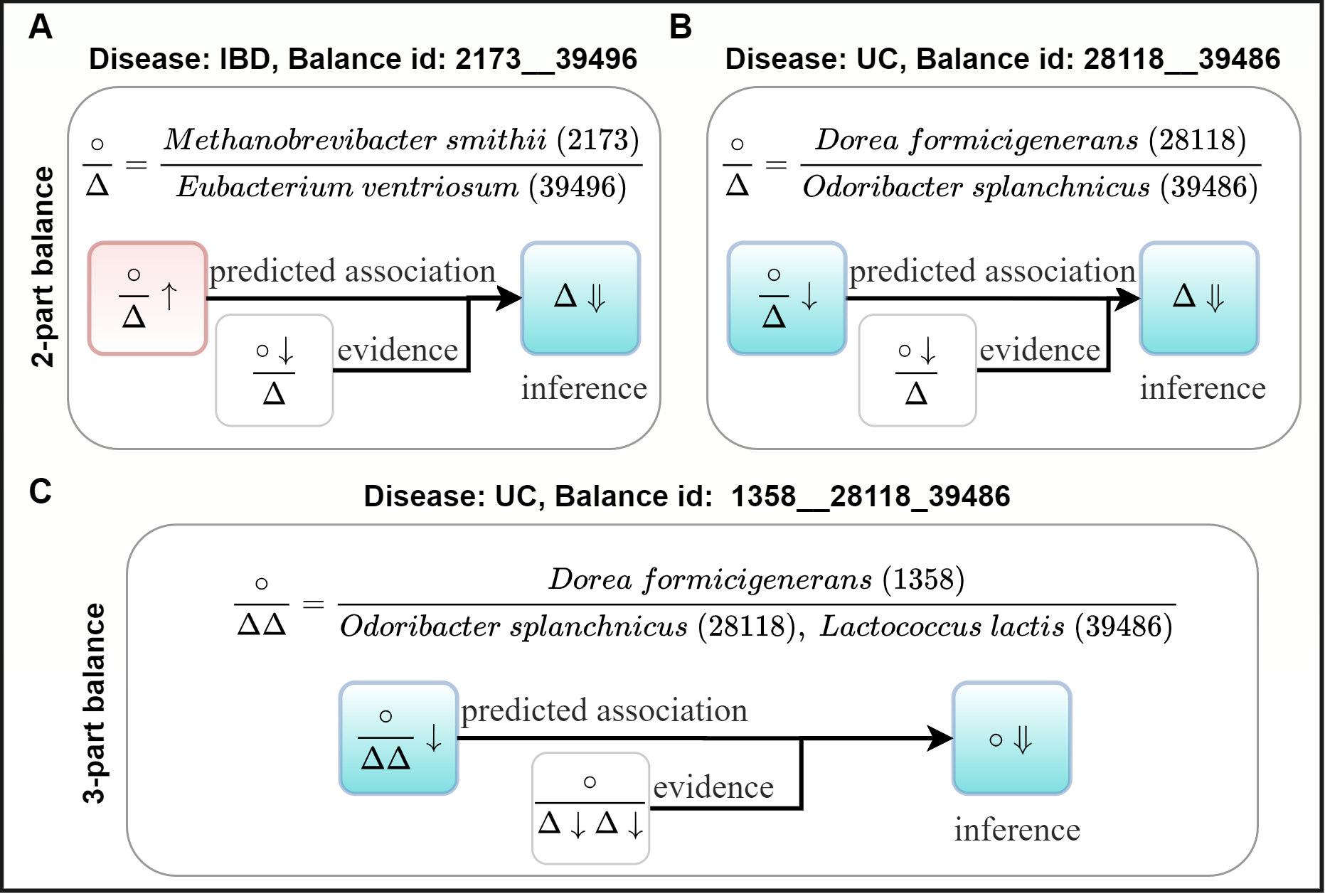
The new taxon-disease associations for IBD and UC datasets can be inferred by combing the predicted balance-disease associations and the evidenced taxon-disease associations in GBDAD.
For the IBD dataset, as illustrated in (A), we inferred that Eubacterium ventriosum is negatively correlated with IBD. This result is supported by a previous study which constructed large-scale personalized microbial community-level metabolic models for IBD patients and healthy controls and discovered that the IBD microbiomes were depleted in contributions of Eubacterium strains such as Eubacterium ventriosum ATCC 27560.
For the UC dataset, as illustrated in (B), we inferred that Odoribacter splanchnicus is negatively correlated with UC, which is consistent with previous discoveries that O. splanchnicus is a known producer of SCFA (Short Chain Fatty Acids, such as acetate, propionate, and butyrate), which can attenuate host inflammation through modulation of immunity. Furthermore, if O. splanchnicus is negatively correlated with UC, we will get another inference that Lactococcus lactis is negatively correlated with UC as well, as illustrated in (C). This result is supported by the fact that the colitis-attenuating effect of L. lactis is well-known, and the prevention and treatment of colitis with L. lactis is widely studied in IBD patients and animal models.
Codes availability
Codes to transform species composition files to balance composition files for each disease are pushed to github (distal_DBA.R.ipynb).
The logistic Regression models were built under the instruction of the mAML automated machine learning pipeline with the Elastic-Net regularization.
Run the code in the notebook (risk_predict.ipynb) to predict disease risks or upload your species composition file to the server to make risk prediction.