Introduction
Identifying key taxonomic biomarkers and restoring commensal gut microbiota is vital for the precision diagnosis and treatment of many human diseases. DisBalance was developed to address the key issues related to microbiome-based binary classifications.
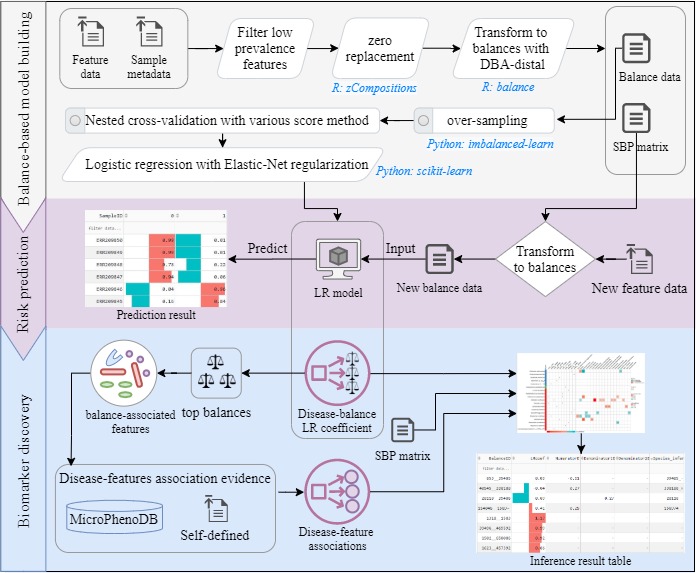
- The distal DBA (distal discriminative balance analysis) method was embedded to select a set of distal discriminative balances as input features for regularized LR (logistic regression) modelling.
- 2111 gut-distributed disease-taxon associations were abstracted from MicroPhenoDB as built-in evidence.
- Multiple strategies for the mining of microbial biomarkers were supplied to facilitate the model-driven and knowledge-driven discoveries.
The implementation of DisBalance is showcased by a complete analysis of a UC demo dataset from GMrepo as demonstrated on the "demo" tab.