Why Balance?
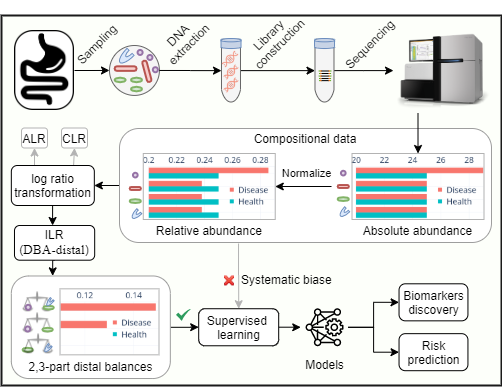
Microbiome data include but are not limited to taxa, genes quantified by high-throughput sequencing (HTS), and proteins, lipids, or metabolites quantified by liquid chromatography-mass spectrometry (LC-MS). HTS and LC-MS both sample abundances of these molecules from the original ecosystems, however, the total number of molecules recorded for each sample is constrained and arbitrary, making these data compositional. Microbiota data are typically interpreted as proportions that add to one. The change of a single microbe will also change the proportions of the remaining microbes. Thus, precisely quantify the changes of the microbes requiring the proper use of mathematical theory.
Balance, a special kind of isometric log-ratio (ILR) transformation applied to the compositional data for various statistical analyses, was introduced by Egozcue and Pawlowsky-Glahn. The distal DBA (discriminative balance analysis) method used in this study selects the distal 2-part and 3-part balances representing log-ratios of 2 or 3 species as features for disease classification. The method outperforms the other strategies in terms of interpretability, runtime, and classification accuracy for the microbiome-based classification tasks.
What is GutBalance?
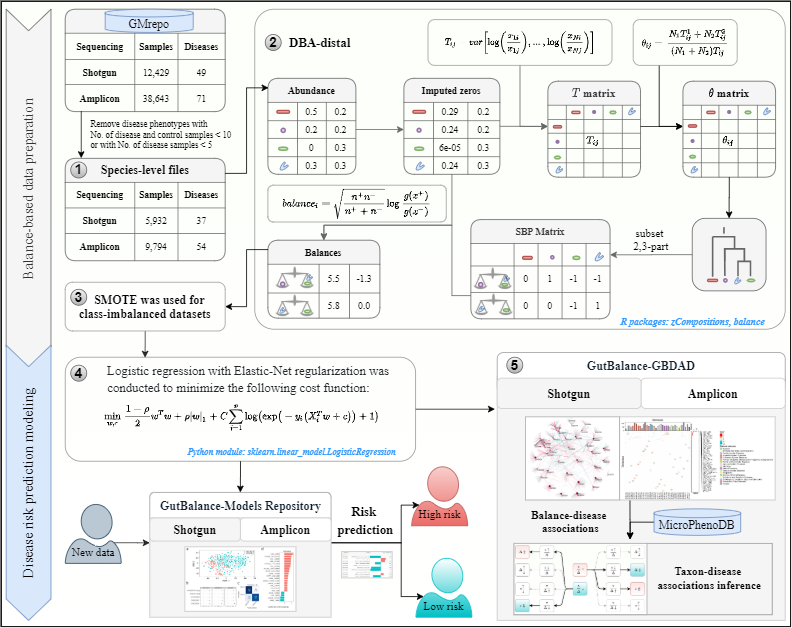
GutBalance is a server for the human gut microbiome-related disease prediction and biomarker discovery based on balances transformed from human gut microbiota species-level relative abundance data (1) retrieved from the GMrepo database.
The DBA-distal balances (2) were abstracted from the relative abundances profile for each disease phenotype by constructing the T matrix, θ matrix and SBP matix. The balances matrix was then constructed by applying the ilr transformation to each balance in the SBP matrix of the respective disease. Third, SMOTE (3) was used to address the imbalanced datasets. Fourth, the prepared feature data were trained with regularized logistic regression (4) and the optimized classifier was obtained for each disease. These classifiers constituted the model repository and support disease risks predition of every new sample. Finally, the predicted balances for each disease classifer were collected to form the GBDAD database (5) with balances-related species linked to the microbes in the MicroPhenoDB database. By combining the balance-disease associations and taxon-disease associations in GBDAD, the new taxon-disease associations may be inferred from the single features aspect.
Cite us
Fenglong Yang, Quan Zou*,
mAML: an automated machine learning pipeline with a microbiome repository for human disease classification,
Database, Volume 2020, 2020, baaa050,
https://doi.org/10.1093/database/baaa050. (SCI, IF2018=3.683)
Fenglong Yang, Quan Zou* and Bo Gao*
GutBalance: a server for the human gut microbiome-based disease prediction and biomarker discovery with compositionality addressed,
Briefings in Bioinformatics, 00(00), 2021, 1–12, bbaa436,
https://doi.org/10.1093/bib/bbaa436. (SCI, IF2020=8.990)
Contact us
For technical problems using GutBalance, please refer to the GutBalance tutorial.
Feel free to report any issues on
GitHub or contact us:
Yang Fenglong (yangfenglong110 ANTI_SPAM_@126.com)
Prof. Quan Zou (zouquan ANTI_SPAM_@nclab.net)